Associação Portuguesa de Investigação em Cancro
New bioimaging approach to quantify protein expression in heterogeneous cancer cell populations
New bioimaging approach to quantify protein expression in heterogeneous cancer cell populations
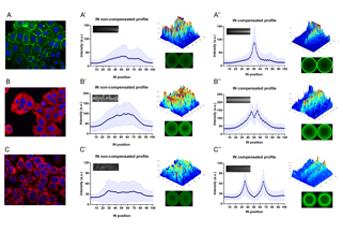
Immunofluorescence microscopy (IF) is a widely used technique that takes advantage of fluorescent-labelled markers to visualize the distribution of molecules at the cellular or at the tissue level. Still, quantitative analysis of fluorescence microscopy images is difficult when dealing with a heterogeneous cell population. The high variability of the expression profiles between cells impairs the extraction of a representative overview/map of a particular target.
In this work, the researchers developed a new analytical tool specific for in situ IF images of heterogeneous cell populations based on a geometric compensation process, which is a classical concept of image registration and alignment. This process is able to deal with cell shape and size variability allowing the generation of a detailed quantitative report and a faithful representation of a large panel of molecules, distributed in distinct cellular compartments.
To experimentally validate the analytical pipeline, the team used synthetic images and real in situ IF images of cell cultures stained for E-cadherin, tubulin and a mitochondria dye, which are the prototypes of membrane, cytoplasmic and organelle-specific markers. The results demonstrated that quantification of protein expression patterns in heterogeneous samples is possible and the method can immediately be translated to research and routine labs.
Authors and affiliations:
Joana Figueiredo1,2, Isabel Rodrigues3, João Ribeiro3, Maria Sofia Fernandes1,2,3, Soraia Melo1,2,4, Bárbara Sousa1,2, Joana Paredes1,2,4, Raquel Seruca1,2,4*, João M. Sanches3*
1 Instituto de Investigação e Inovação em Saúde (i3S), Porto, Portugal;
2 Institute of Molecular Pathology and Immunology of the University of Porto (IPATIMUP), Porto, Portugal;
3 Institute for Systems and Robotics (ISR/IST), LARSyS, Bioengineering
Dept, Instituto Superior Técnico, Universidade de Lisboa, Lisbon, Portugal;
4 Medical Faculty of the University of Porto, Porto, Portugal.
Abstract:
Immunofluorescence is the gold standard technique to determine the level and spatial distribution of fluorescent-tagged molecules. However, quantitative analysis of fluorescence microscopy images faces crucial challenges such as morphologic variability within cells. In this work, we developed an analytical strategy to deal with cell shape and size variability that is based on an elastic geometric alignment algorithm. Firstly, synthetic images mimicking cell populations with morphological variability were used to test and optimize the algorithm, under controlled conditions. We have computed expression profiles specifically assessing cell-cell interactions (IN profiles) and profiles focusing on the distribution of a marker throughout the intracellular space of single cells (RD profiles). To experimentally validate our analytical pipeline, we have used real images of cell cultures stained for E-cadherin, tubulin and a mitochondria dye, selected as prototypes of membrane, cytoplasmic and organelle-specific markers. The results demonstrated that our algorithm is able to generate a detailed quantitative report and a faithful representation of a large panel of molecules, distributed in distinct cellular compartments, independently of cell’s morphological features. This is a simple end-user method that can be widely explored in research and diagnostic labs to unravel protein regulation mechanisms or identify protein expression patterns associated with disease.
Journal: Scientific Reports
Link: https://www.nature.com/articles/s41598-018-28570-z




