Associação Portuguesa de Investigação em Cancro
i3S lidera estudo que redefine o risco de cancro e as diretrizes de testes para portadores do gene CTNNA1
i3S lidera estudo que redefine o risco de cancro e as diretrizes de testes para portadores do gene CTNNA1
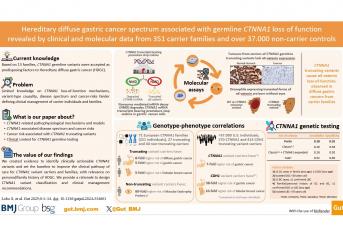
Um estudo internacional envolvendo 56 investigadores e clínicos de nove países, liderado pela cientista Carla Oliveira, do Instituto de Investigação e Inovação em Saúde da Universidade do Porto (i3S), e publicado na prestigiada revista GUT, definiu novas causas de risco genético de cancro gástrico e mamário e critérios clínicos para os identificar. Os já chamados «Critérios do Porto» abrem caminho para melhorias nos testes genéticos e na gestão clínica de indivíduos afetados com cancro gástrico difuso hereditário, nomeadamente na definição de medidas de vigilância e/ou de prevenção.
Este estudo foca-se no papel das variantes hereditárias de um determinado gene, o CTNNA1, no cancro gástrico difuso hereditário (HDGC). A equipa analisou dados de 1.308 indivíduos de 351 famílias portadoras de variantes neste gene, em comparação com 37.428 não-portadores, maioritariamente de ascendência europeia e americana. «Utilizando uma combinação de análises de dados clínicos e moleculares — incluindo edição genética avançada com CRISPR/Cas9 em modelos celulares e animais — e integração com dados populacionais, descobrimos como as variantes no CTNNA1 promovem o desenvolvimento de cancro e que tipos de cancro surgem em portadores destas variantes», explica Silvana Lobo, primeira autora do artigo.
A equipa, acrescenta a investigadora, «descobriu um mecanismo molecular que causa a perda de expressão do gene CTNNA1, essencial para a integridade do tecido gástrico, e que portadores de variantes inactivantes têm um risco de desenvolver cancro sete vezes superior do que não portadores, e oito vezes superior do que portadores de outro tipo de variantes no gene CTNNA1». Os investigadores demonstraram também que, apesar de portadores de variantes hereditárias no gene CTNNA1 terem risco de doença aumentado, este é significativamente mais baixo quando comparado com os portadores de variantes hereditárias no gene CDH1.
<«Observámos igualmente que em famílias onde existem casos de cancro gástrico, os portadores de variantes hereditárias do CTNNA1 desenvolvem, com frequência, cancro da mama lobular e que o cancro da mama é particularmente frequente nas famílias norte-americanas, enquanto as famílias europeias apresentam preferencialmente cancro gástrico difuso», adianta Carla Oliveira, líder do grupo do i3S «Expression Regulation in Cancer».
Este novo conjunto de diretrizes, que os investigadores chamaram de “Critérios do Porto”, sublinha Carla Oliveira, «propõe novos critérios para seleção de famílias suspeitas para realização de teste genético, baseados em dados clínicos reais, e por isso, aumenta em 9% a deteção de famílias com variantes no CTNNA1, sem comprometer a precisão». O estudo, acrescenta a investigadora, «salienta ainda a importância de conhecer a história clínica das famílias e dirigir o teste genético do CTNNA1 às famílias que cumpram os critérios clínicos adequados».
Autores e Afiliações:
Silvana Lobo1,2, Alexandre Dias1,2, Ana Maria Pedro1, Marta Ferreira1,3, André Pinto-Oliveira1, Celina São José1, Jennifer Herrera-Mullar4, Nádia Pinto1,52,53, Chrystelle Colas5,6, Robert Hüneburg7,8, Jacob Nattermann7,8, Lise Boussemart9, Liselotte P van Hest10, Leticia Moreira11,12, Carolyn Horton4, Dana Farengo Clark13, Sigrid Tinschert14, Lisa Golmard5, Isabel Spier15,8, Adrià López-Fernández16, Daniela Oliveira17, Magali Svrcek18, Pierre Bourgoin19, Florence Coulet20, Hélène Delhomelle5,21, Jeremy Davis22, Birthe Zäncker23, Conxi Lazaro24,25, Joana Guerra26,27,28, Maria L. Almeida29, Sergio Carrera30, Ana Patiño31,32,33, Paul Gundlach34, Monika Laszkowska35,36, Vivian E. Strong37, Manuel R. Teixeira26,28,2, Intan Schrader38, Verena Steinke-Lange39, Irene Gullo1,40,41, Sérgio Sousa17, Manuela Batista42, Stefan Aretz15,8, Judith Balmaña16,43, Melyssa Aronson44, Augusto Perazzolo Antoniazzi45, Edenir I Palmero46,47, Paul Mansfield48, Lizet E van der Kolk49, Annemieke Cats50, Jolanda M van Dieren50, Sergi Castellví-Bel11, Bryson Katona13, Rachid Karam4, Paulo S. Pereira51,1, Patrick R. Benusiglio20, Carla Oliveira52,1,41,*
1i3S - Instituto de Investigação e Inovação em Saúde, University of Porto, Portugal
2ICBAS -School of Medicine and Biomedical Sciences of the University of Porto, Portugal
3Department of Computer Science, Faculty of Science, University of Porto, Rua do Campo Alegre s/n, 4169-007 Porto, Portugal
4AmbryGenetics, California, United States
5Department of genetics, Curie Institute, Paris, France
6INSERM U830, Université de Paris, Paris, France
7Department of Internal Medicine I, University Hospital Bonn, Germany
8National Center for Hereditary Tumor Syndromes, University Hospital Bonn, Germany
9Nantes Université, Univ Angers, CHU Nantes, INSERM, Immunology and New Concepts in ImmunoTherapy, France
10Department of Human Genetics, Section Clinical Genetics, Amsterdam UMC, location University of Amsterdam, Amsterdam, the Netherlands
11Department of Gastroenterology, Hospital Clínic de Barcelona; Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS); Centro de Investigación Biomédica en Red de Enfermedades Hepáticas y Digestivas (CIBEREHD)
12Facultat de Medicina i Ciències de la Salud, Universitat de Barcelona (UB), Barcelona, Spain
13University of Pennsylvania Perelman School of Medicine, Division of Gastroenterology and Hepatology, Pennsylvania, USA
14Medical University Innsbruck, Division of Human Genetics, Austria
15Institute of Human Genetics, Medical Faculty, University of Bonn, Bonn, Germany
16Hereditary Cancer Genetics Group, Medical Oncology Department, Hospital Vall d'Hebron and Vall d'Hebron Institute of Oncology, Barcelona, Spain
17Paediatric Hospital, Coimbra Hospital and University Centre, Medical Genetics Unit, Portugal
18Sorbonne Université, Equipe Instabilité Des Microsatellites Et Cancer, Centre de Recherche Saint Antoine, France
19Sorbonne Université, Laboratoire D'anatomie Et Cytologie Pathologiques, Hôpital Saint-Antoine, France
20Sorbonne Université, Hôpital Pitié-Salpêtrière, Département de Génétique médicale, APHP, Paris, France
21Paris Sciences & Lettres Research University, Paris, France
22Center for Cancer Research, National Cancer Institute, Maryland, USA
23Institut für Klinische Genetik, Universitätsklinikum CarlGustav Carus Dresden, Germany
24Hereditary Cancer Program, Molecular Mechanisms and Experimental Therapy in Oncology (Oncobell) Program, Catalan Institute of Oncology, Institut d’Investigació Biomèdica de Bellvitge (IDIBELL), l’Hospitalet del Llobregat, Spain
25Centro de Investigación Biomédica en Red de Cáncer (CIBERONC), Spain
26Cancer Genetics Group, Research Center of IPO Porto (CI-IPOP)/CI-IPOP@RISE (Health Research Network), Portuguese Oncology Institute of Porto (IPO-Porto)/Porto Comprehensive Cancer Center (Porto.CCC), Porto, Portugal
27Doctoral Programme in Biomedical Sciences, School Medicine and Biomedical Sciences, University of Porto (ICBAS-UP), Porto, Portugal
28Department of Laboratory Genetics, Portuguese Oncology Institute of Porto (IPO-Porto)/Porto Comprehensive Cancer Center (Porto.CCC), Porto, Portugal
29ULSBraga – Unidade de Saúde Local de Braga, Braga, Portugal
30Genetic Counseling Unit-Medical Oncology Department, Cruces University Hospital, Barakaldo. Spain
31Medical Genomics Unit, Cancer Center, University Clinic of Navarra, Spain
32Laboratory of Advanced Therapies for Pediatric Solid Tumors, Cancer Division, Cima, Spain
33A701 - Advanced Therapies for Pediatric Solid Tumors, IdiSNA, Spain
34Department of Clinical Genetics, Erasmus MC Cancer Institute, University Medical Center, Rotterdam, the Netherlands
35Gastroenterology, Hepatology, and Nutrition Service, Department of Subspecialty Medicine, Memorial Sloan Kettering Cancer Center
36Weill Cornell Medical School of Cornell University
37Memorial Sloan Kettering Cancer Center, New York, USA
38Hereditary Cancer Program, BC Cancer, Vancouver, Canada
39Medizinische Klinik und Poliklinik IV, Klinikum der Universität München, Germany
40Department of Pathology, Unidade Local de Saúde de São João, Porto, Portugal
41FMUP - Faculty of Medicine of the University of Porto, Portugal
42Serviço de Cirurgia Geral, Unidade Local de Saúde de São João, Porto, Portugal
43Universitat Autònoma de Barcelona, 08193 Bellaterra, Spain
44Zane Cohen Centre, Sinai Health System, Toronto, Canada
45Barretos Cancer Hospital, Cancer Genetics Departament, Brazil
46Molecular Oncology Research Center, Barretos Cancer Hospital, Brazil
47Department of Genetics, Brazilian National Cancer Institute, Rio de Janeiro, Brazil
48The University of Texas MD Anderson Cancer Center, Texas, USA
49Department of Clinical Genetics, the Netherlands Cancer Institute, The Netherlands
50Department of Gastrointestinal Oncology, the Netherlands Cancer Institute, Amsterdam, the Netherlands
51IBMC - Instituto de Biologia Molecular e Celular, University of Porto, Portugal
52IPATIMUP - Institute of Pathology and Molecular Immunology of the University of Porto, Portugal
53Center of Mathematics, University of Porto, Porto, Portugal
*Corresponding author ([email protected])
Abstract:
Background: Diffuse gastric cancer (DGC) is the most common manifestation in germline CTNNA1 variant carriers, with one study estimating a 49–57% lifetime risk by age 80. Knowledge on CTNNA1-associated hereditary diffuse gastric cancer (HDGC), loss-of-function mechanisms, variant-type causality, disease spectrum and cancer risks remains scarce.
Objective: Explore CTNNA1 genotype–phenotype associations to improve genetic testing criteria, surveillance and risk-reduction recommendations for carriers.
Design: Using molecular, clinical and population data from 1308 individuals from 351 CTNNA1-variant carrier families and 37 428 non-carriers from European and American ancestries, we analysed genotype–phenotype associations with multivariable logistic regression. With CRISPR/Cas9 CTNNA1-knockout gastric cancer (GC) cells and CTNNA1-humanised Drosophila, we assessed CTNNA1-associated loss-of-function mechanisms.
Results: CTNNA1-truncating transcripts are degraded by nonsense-mediated mRNA decay (NMD), and DGCs from germline CTNNA1-truncating carriers lose αE-catenin. These transcripts are non-functional in Drosophila, in contrast to non-truncating transcripts. DGC risk is eightfold higher in truncating, compared with non-truncating carriers. The risk of GC and lobular breast cancer (LBC) development in CTNNA1-truncating variant carriers is fivefold and eightfold lower than in CDH1 pathogenic/likely pathogenic variant carriers, respectively. Compared with wild-type individuals, GC risk is 7-fold higher in CTNNA1-truncating and 38-fold higher in CDH1-truncating variant carriers. LBC is recurrent among CTNNA1-truncating carriers, some lacking HDGC criteria. Simplification of previous criteria for CTNNA1 genetic testing produced the ‘Porto’ criteria, which increased CTNNA1-carrier families’ pick-up rate by 9%, without performance loss compared with the HDGC 2020 clinical guidelines. Macular dystrophy patterned-2 was positively associated with non-truncating variants, specifically in the αE-catenin M-fragment.
Conclusion: We provide compelling evidence supporting that CTNNA1-truncating variants positively associate with DGC and LBC, and NMD as the pathophysiological mechanism leading to CTNNA1 downregulation. We demonstrate that compared with CDH1, CTNNA1 is a moderate penetrance HDGC gene. This new knowledge is essential to define surveillance and/or prophylactic measures for CTNNA1-carrier individuals and families.
Revista: GUT
Link




