Associação Portuguesa de Investigação em Cancro
Biomedical implications of protein synthesis errors
Biomedical implications of protein synthesis errors
Authors and Affiliations:
Marisa Reverendo1, Ana R Soares1, Patrícia M Pereira1, Laura Carreto1, Violeta Ferreira1, Evelina Gatti 2, 3, 4, Philippe Pierre 2, 3, 4, Gabriela R Moura1 and Manuel A S Santos1*
1iBiMED & Health Sciences, University of Aveiro, Portugal
2 Centre d’Immunologie de Marseille-Luminy,
Aix-Marseille Université, Marseille, France
3 INSERM, Marseille, France
4 CNRS, Marseille, France
Abstract:
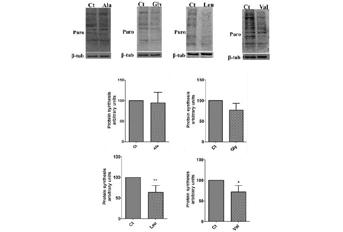
Mutations in genes that encode tRNAs, aminoacyl-tRNA syntheases, tRNA modifying enzymes and other tRNA interacting partners are associated with neuropathies, cancer, type-II diabetes and hearing loss, but how these mutations cause disease is unclear. We have hypothesized that levels of tRNA decoding error (mistranslation) that do not fully impair embryonic development can accelerate cell degeneration through proteome instability and saturation of the proteostasis network. To test this hypothesis we have induced mistranslation in zebrafish embryos using mutant tRNAs that misincorporate Serine (Ser) at various non-cognate codon sites. Embryo viability was affected and malformations were observed, but a significant proportion of embryos survived by activating the unfolded protein response (UPR), the ubiquitin proteasome pathway (UPP) and downregulating protein biosynthesis. Accumulation of reactive oxygen species (ROS), mitochondrial and nuclear DNA damage and disruption of the mitochondrial network, were also observed, suggesting that mistranslation had a strong negative impact on protein synthesis rate, ER and mitochondrial homeostasis. We postulate that mistranslation promotes gradual cellular degeneration and disease through protein aggregation, mitochondrial dysfunction and genome instability.
Journal: RNA Biology
Link: https://www.landesbioscience.com/journals/rnabiology/article/32199/?nocache=190466892




