Associação Portuguesa de Investigação em Cancro
Nova função da Histona Hist2h2ac na regulação da proliferação e migração das células do cancro da mama
Nova função da Histona Hist2h2ac na regulação da proliferação e migração das células do cancro da mama
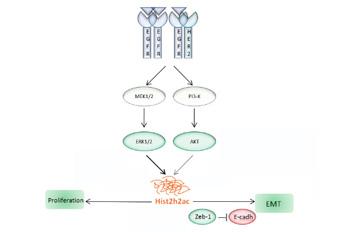
Proliferation and differentiation are controlled through chromatin remodelling. Therefore, there is an enormous biological significance and clinical value in understanding how specific signalling pathways are affected by histone replacement in the nucleosome. In this work, mass spectrometry was used to screen HC11 mammary epithelial cells for changes in histone levels throughout cell differentiation. The canonical histone isoform Histone H2A type 2-C (Hist2h2ac) was found only in undifferentiated/proliferating cells. Hist2h2ac mRNA was induced by EGF, specifically in the CD24+/CD29hi/DC44hi cell subpopulation. Hist2h2ac mRNA was increased by MEK1/2 or PI3-K activation in HC11 and EpH4 mammary epithelial cells, and in MC4-L2 and T47-D breast cancer cells. Hist2h2ac silencing inhibited EGF-induced Zeb-1 expression and E-cadherin down-regulation, and this effect was reverted by Hist2h2ac re-expression. Notably, silencing of Hist2h2ac increased EGFR, ERBB2, and ERK1/2 activation but did not allow EGF-induced proliferation. HIST2H2AC was expressed in all breast cancer molecular subtypes and found altered in 17% breast cancers, being 16.8% of the cases related to HIST2H2AC gene amplification and/or mRNA upregulation. In summary, this is the first study that identifies a canonical histone isoform -Hist2h2ac-downstream of the EGFR pathway, regulating oncogenic signalling and thereby contributing to deregulation of target genes.
Fátima Liliana Monteiro (a, b), Rui Vitorino (a, c), Jun Wang (d), Hugo Cardoso (a), Hugo Laranjeira (a), Joana Simões (a), Margarida Caldas (e), Rui Henriqueb (e, f), Francisco Amado (a), Cecilia Williams (d, g, h), Carmen Jerónimo (b, f), Luisa A. Helguero (a, c)
a) Mass Spectrometry Centre, Organic Chemistry and Natural Products Unit, Dep. of Chemistry, Universidade de Aveiro, Campus de Santiago, 3810-193, Aveiro, Portugal
b) Cancer Biology and Epigenetics Group, Portuguese Oncology Institute of Porto, 4200-072 Porto, Portugal
c) Institute for Biomedicine -iBiMED, Department of Medical Sciences, University of Aveiro, Portugal
d) Department of Biology and Biochemistry, Center for Nuclear Receptors and Cell Signalling, University of Houston, Houston, TX 77204, USA
e) Department of Pathology, Portuguese Oncology Institute of Porto, 4200-072 Porto, Portugal
f) Department of Pathology and Molecular Immunology, Institute of Biomedical Sciences Abel Salazar (ICBAS) – University of Porto, 4050-313 Porto, Portugal
g) Science for Life Laboratory, Division of Proteomics, School of Biotechnology, KTH - Royal Institute of Technology, 171 21 Solna, Sweden
h) Department of Biosciences, Karolinska Institutet, Huddinge, Sweden
Summary Alterations in the way the genetic information is packed in the cell nucleus determines cell fate (differentiation stage) and when disrupted can result in cancer. DNA containing the genetic information for complete cellular differentiation and function is packed within specialized proteins known as histones. There are 5 histone families H1, H2A, H2B, H3 and H4 Until recently it was thought that most canonical histones within a family were interchangeable and shared the same function. Our work identified changes in histone patterns according to the cell differentiation stage. We showed that a specific histone (Hist2h2ac) is present in high quantities in cells with high proliferation rate and low differentiation state. In addition, Hist2h2ac is needed to activate cell division and migration in breast cancer cell lines. We further confirmed that Hist2h2ac is present in human breast cancer and its gene is mutated or overexpressed in 16% cases. In summary, this work showed that histones have a specific expression pattern according to the cell differentiation stage. Moreover, we identified a new protein which is necessary to induce genetic programs highly active in cancer.
Cancer Letters
http://www.sciencedirect.com/science/article/pii/S0304383517301714




